Principal Component Analysis (PCA) is a technology for simplified analysis of data sets. PCA uses variance decomposition to reduce the dimensionality of multi-dimensional data, remove noise and redundancy, and reveal the most important elements and structures hidden behind complex data. In the field of life sciences, in proteomics and metabolomics research applications, it is usually necessary to observe data containing multiple variables and collect a large amount of data to analyze and find rules. In addition, the microbial community structure is affected by many factors, such as light, temperature, humidity, etc. It is necessary to understand whether the purpose grouping is related to a certain factor. CD Genomics often applies the PCA sorting method to visually analyze the data.
Principal Component Analysis Method
The commonly used tools in PCA are the PCA module in GCTA, Canoco software, smartpca in EIGENSOFT, and many R packages that can do PCA analysis. The visualization operation is generally implemented using the ggplot package following the analysis.

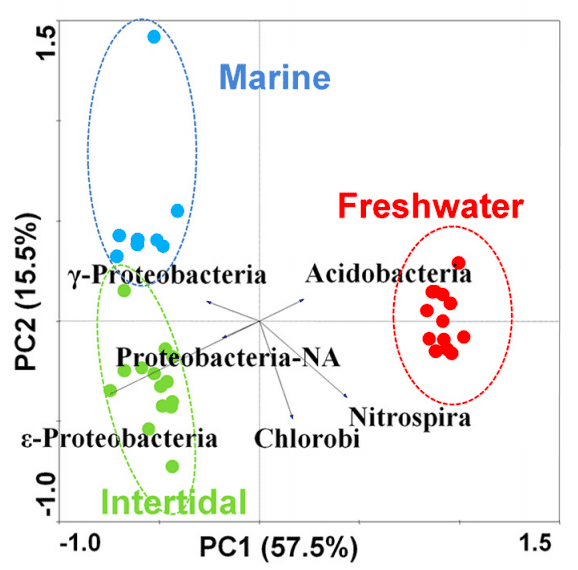
Fig 1. Principal component analysis (PCA) of phylum abundance data using Canoco 4.5. (Wang Y, et al. 2012)
Different colors or shapes in the PCA diagram represent sample groups under different environments or conditions. The scales on the horizontal and vertical axes are relative distances and have no practical meaning. Principal component 1 (PC1) and principal component 2 (PC2) represent the suspected influencing factors of the deviation of the three groups of sample composition, which need to be summarized in combination with the sample feature information. The contribution rates of PC1 and PC2 are 57.7% and 15.5%, respectively.
learn more: atac sequencing
Advantages of Principal Component Analysis
PCA can solve three major problems faced by big data analysis:
High-dimensional problems by reducing dimensionality.
Reducing the dimensionality can effectively remove redundant data and ensure that the loss of feature information is minimized.
The data after dimensionality reduction can be displayed visually to facilitate the interpretation of effective information in big data.
Application Field
Microbial principal component analysis.
Proteomics analysis.
Metabolomics analysis.
PCA is a common biological information analysis. A biological information service provider, CD Genomics can provide PCA services. Once we receive the data to be analyzed, we will analyze them and provide a detailed analysis report along with professional charts. If you have any questions or other analysis needs, please feel free to contact us.
Reference
- Wang Y, et al. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags.[J]. Applied & Environmental Microbiology, 2012, 78(23):8264.